Titanic Kaggle challenge
Vincent Brouté • March 8, 2018
dataHere is my first kernel along with my very first attempt with the Titanic challenge (newbie still learning ...). The goal is to achieve a binary classification in order to predict whether a passenger survived the Titanic sinking (variable 'Survived'). Read on Kaggle.
Data import & wrangling
Titanic training dataset overview
| PassengerId | Survived | Pclass | Name | Sex | Age | SibSp | Parch | Ticket | Fare | Cabin | Embarked |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 0 | 3 | Braund, Mr. Owen Harris | male | 22 | 1 | 0 | A/5 21171 | 7.2500 | NA | S |
| 2 | 1 | 1 | Cumings, Mrs. John Bradley (Florence Briggs Thayer) | female | 38 | 1 | 0 | PC 17599 | 71.2833 | C85 | C |
| 3 | 1 | 3 | Heikkinen, Miss. Laina | female | 26 | 0 | 0 | STON/O2. 3101282 | 7.9250 | NA | S |
| 4 | 1 | 1 | Futrelle, Mrs. Jacques Heath (Lily May Peel) | female | 35 | 1 | 0 | 113803 | 53.1000 | C123 | S |
| 5 | 0 | 3 | Allen, Mr. William Henry | male | 35 | 0 | 0 | 373450 | 8.0500 | NA | S |
| 6 | 0 | 3 | Moran, Mr. James | male | NA | 0 | 0 | 330877 | 8.4583 | NA | Q |
| 7 | 0 | 1 | McCarthy, Mr. Timothy J | male | 54 | 0 | 0 | 17463 | 51.8625 | E46 | S |
| 8 | 0 | 3 | Palsson, Master. Gosta Leonard | male | 2 | 3 | 1 | 349909 | 21.0750 | NA | S |
| 9 | 1 | 3 | Johnson, Mrs. Oscar W (Elisabeth Vilhelmina Berg) | female | 27 | 0 | 2 | 347742 | 11.1333 | NA | S |
| 10 | 1 | 2 | Nasser, Mrs. Nicholas (Adele Achem) | female | 14 | 1 | 0 | 237736 | 30.0708 | NA | C |
At first sight
- Some variables are qualitative : Pclass (1, 2 or 3), Sex (male or female) and Embarked (C, Q or S).
- Some variables are quantitative : Age, SibSp, Parch and Fare.
- Some variables contain missing values.
- Within the 'Name' variable, the passengers title (Miss., Master. Captain.) could be a usefull information for the predictions.
Missing values rate within variables
map_dbl(titanicTrainingDataset, function(x) mean(is.na(x)))
## PassengerId Survived Pclass Name Sex Age
## 0.000000000 0.000000000 0.000000000 0.000000000 0.000000000 0.198653199
## SibSp Parch Ticket Fare Cabin Embarked
## 0.000000000 0.000000000 0.000000000 0.000000000 0.771043771 0.002244669
The Age variable contains 19.86% of missing values and the Cabin one contains 77.1%. I will not use Cabin variable as there is too many missing values.
Import Titanic training dataset with proper col types for categorical variables
titanicTrainingDataset <- read_csv(
trainFile,
col_types = cols(
Survived = col_factor(0:1),
Pclass = col_factor(1:3),
Sex = col_factor(c("male", "female")),
Embarked = col_factor(c("C", "Q", "S"))
)
) %>%
filter(!is.na(Embarked)) %>%
mutate(Age = ifelse(is.na(Age), median(Age, na.rm = T), Age))
I filled the blank values for the Age variable with the median age and I have excluded rows that have no value for 'Embarked'.
Extract the passengers title from their name into a new dedicated variable
titanicTrainingDataset <- titanicTrainingDataset %>%
mutate(Title = as.factor(str_extract(Name, regex("([a-z]+\\.)", ignore_case = T))))
levels(titanicTrainingDataset$Title)
## [1] "Capt." "Col." "Countess." "Don." "Dr."
## [6] "Jonkheer." "Lady." "Major." "Master." "Miss."
## [11] "Mlle." "Mme." "Mr." "Mrs." "Ms."
## [16] "Rev." "Sir."
17 distinct titles have been extracted from the 'Name' variable.
Exploratory Data Analysis
summary(titanicTrainingDataset)
## PassengerId Survived Pclass Name Sex
## Min. : 1 0:549 1:214 Length:889 male :577
## 1st Qu.:224 1:340 2:184 Class :character female:312
## Median :446 3:491 Mode :character
## Mean :446
## 3rd Qu.:668
## Max. :891
##
## Age SibSp Parch Ticket
## Min. : 0.42 Min. :0.0000 Min. :0.0000 Length:889
## 1st Qu.:22.00 1st Qu.:0.0000 1st Qu.:0.0000 Class :character
## Median :28.00 Median :0.0000 Median :0.0000 Mode :character
## Mean :29.32 Mean :0.5242 Mean :0.3825
## 3rd Qu.:35.00 3rd Qu.:1.0000 3rd Qu.:0.0000
## Max. :80.00 Max. :8.0000 Max. :6.0000
##
## Fare Cabin Embarked Title
## Min. : 0.000 Length:889 C:168 Mr. :517
## 1st Qu.: 7.896 Class :character Q: 77 Miss. :181
## Median : 14.454 Mode :character S:644 Mrs. :124
## Mean : 32.097 Master.: 40
## 3rd Qu.: 31.000 Dr. : 7
## Max. :512.329 Rev. : 6
## (Other): 14
titanicTrainingDataset %>%
ggplot(aes(x = Survived)) +
geom_bar()
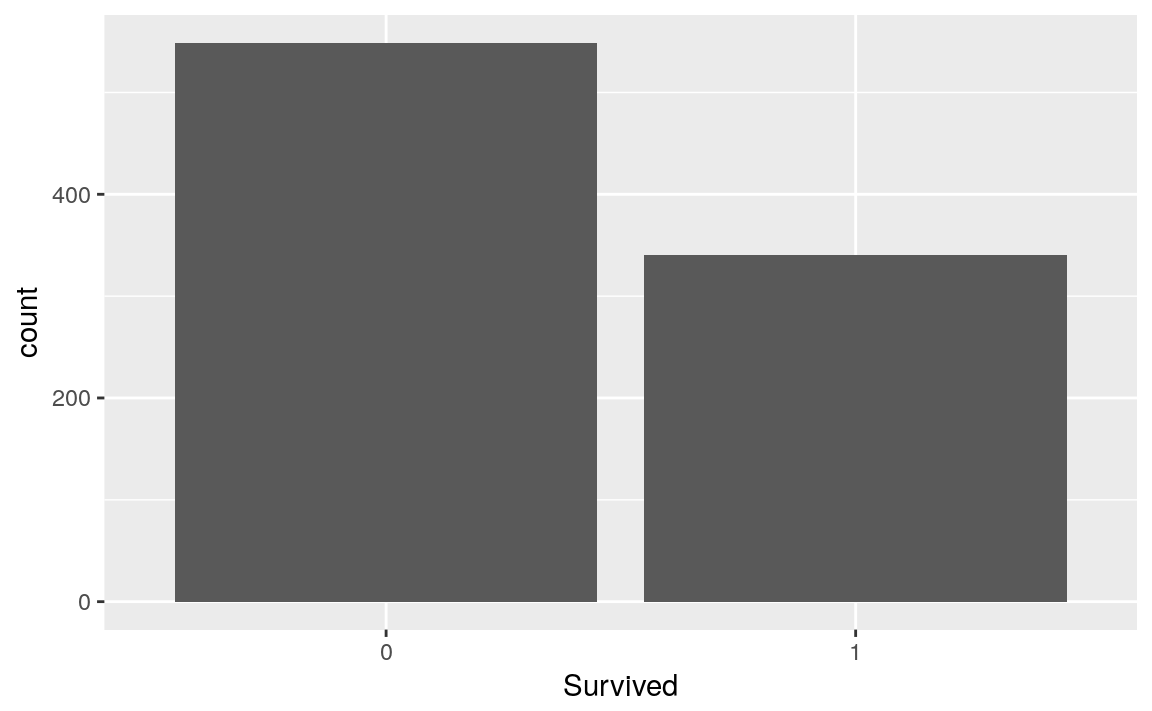
The following function will help me to visualize the response 'Survived' against each qualitative or quantitative potential predictors.
analysePredictorResponse <- function(data, predictor, response) {
if (is.factor(data[[predictor]])) {
ggplot(mapping = aes(data[[predictor]], fill = data[[response]])) +
geom_bar() +
labs(title = paste(predictor, "vs", response), x = predictor, fill = response)
} else {
chart1 <- ggplot(mapping = aes(data[[response]], data[[predictor]])) +
geom_boxplot() +
labs(title = paste(predictor, "vs", response), x = response, y = predictor)
chart2 <- ggplot(mapping = aes(x = data[[predictor]], , y = ..density.., colour = data[[response]])) +
geom_freqpoly(position = "dodge") +
labs(title = paste(predictor, "vs", response), colour = response, x = predictor)
grid.arrange(chart1, chart2)
}
}
Age vs Survived
analysePredictorResponse(titanicTrainingDataset, 'Age', 'Survived')
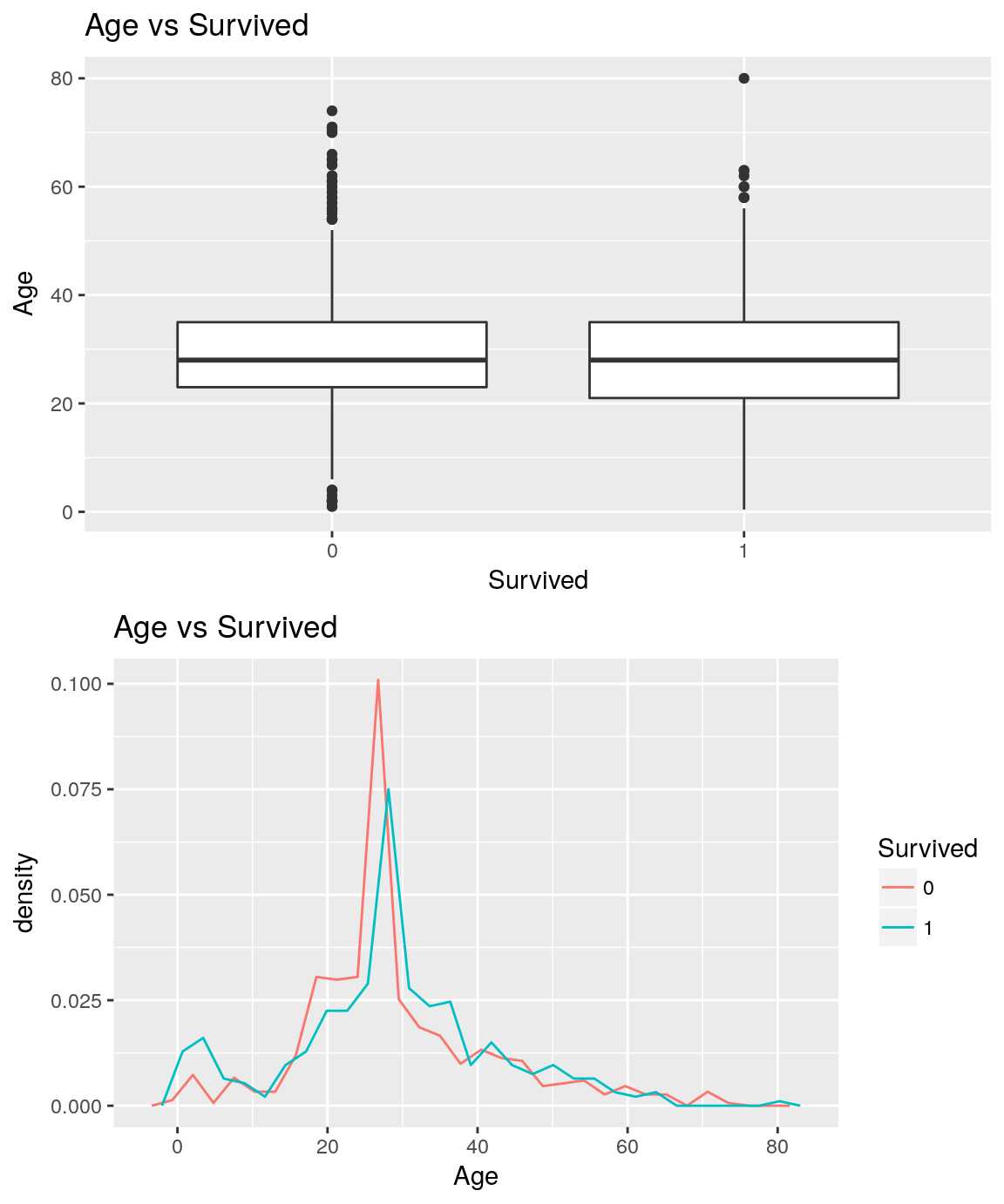
Sex vs Survived
analysePredictorResponse(titanicTrainingDataset, 'Sex', 'Survived')
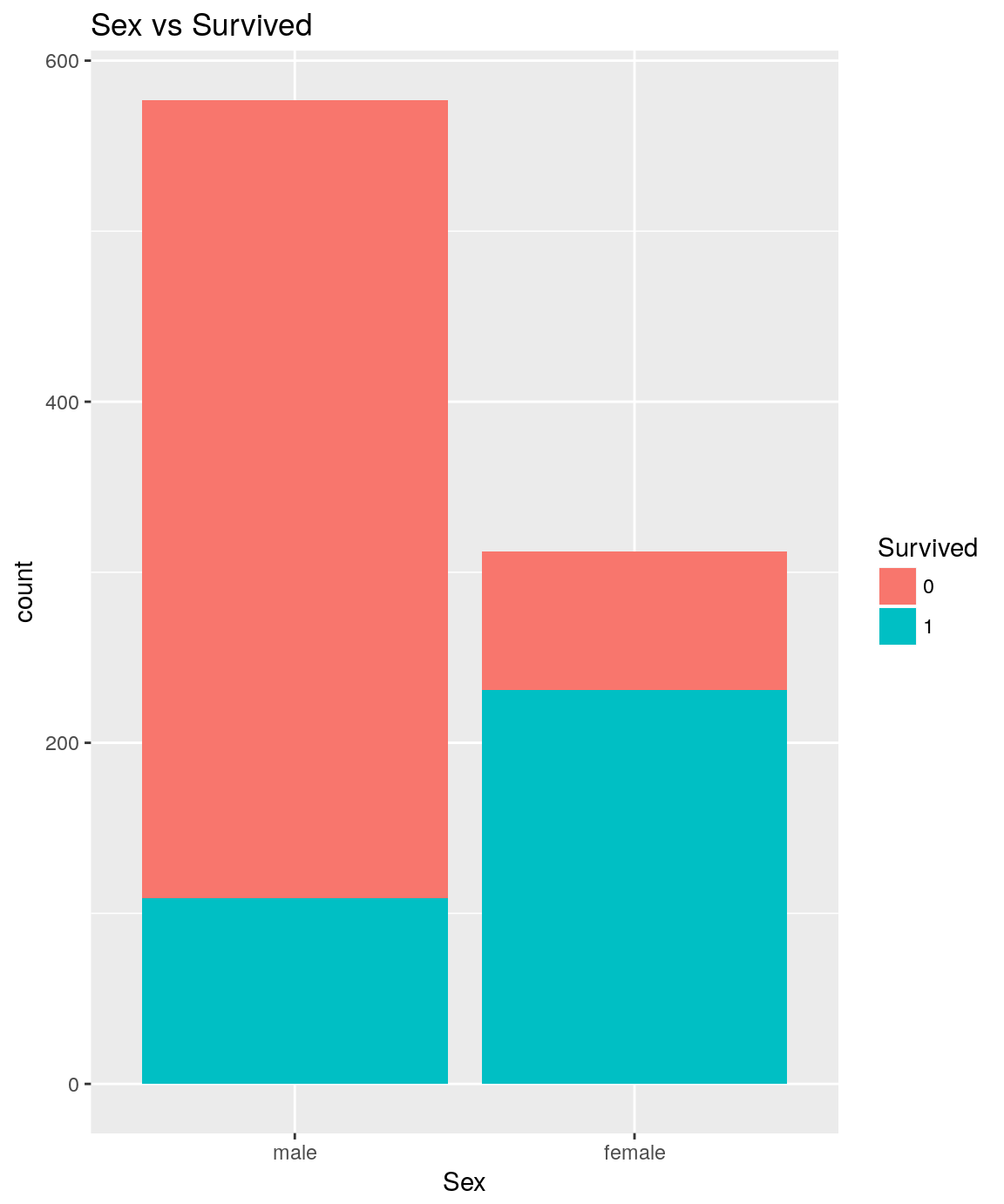
titanicTrainingDataset %>%
group_by(Sex) %>%
summarize(SurvivedRatio = mean(Survived == 1))
## # A tibble: 2 x 2
## Sex SurvivedRatio
## <fct> <dbl>
## 1 male 0.189
## 2 female 0.740
It seems obvious that females have significantly more chance than males to survive.
Pclass vs Survived
analysePredictorResponse(titanicTrainingDataset, 'Pclass', 'Survived')
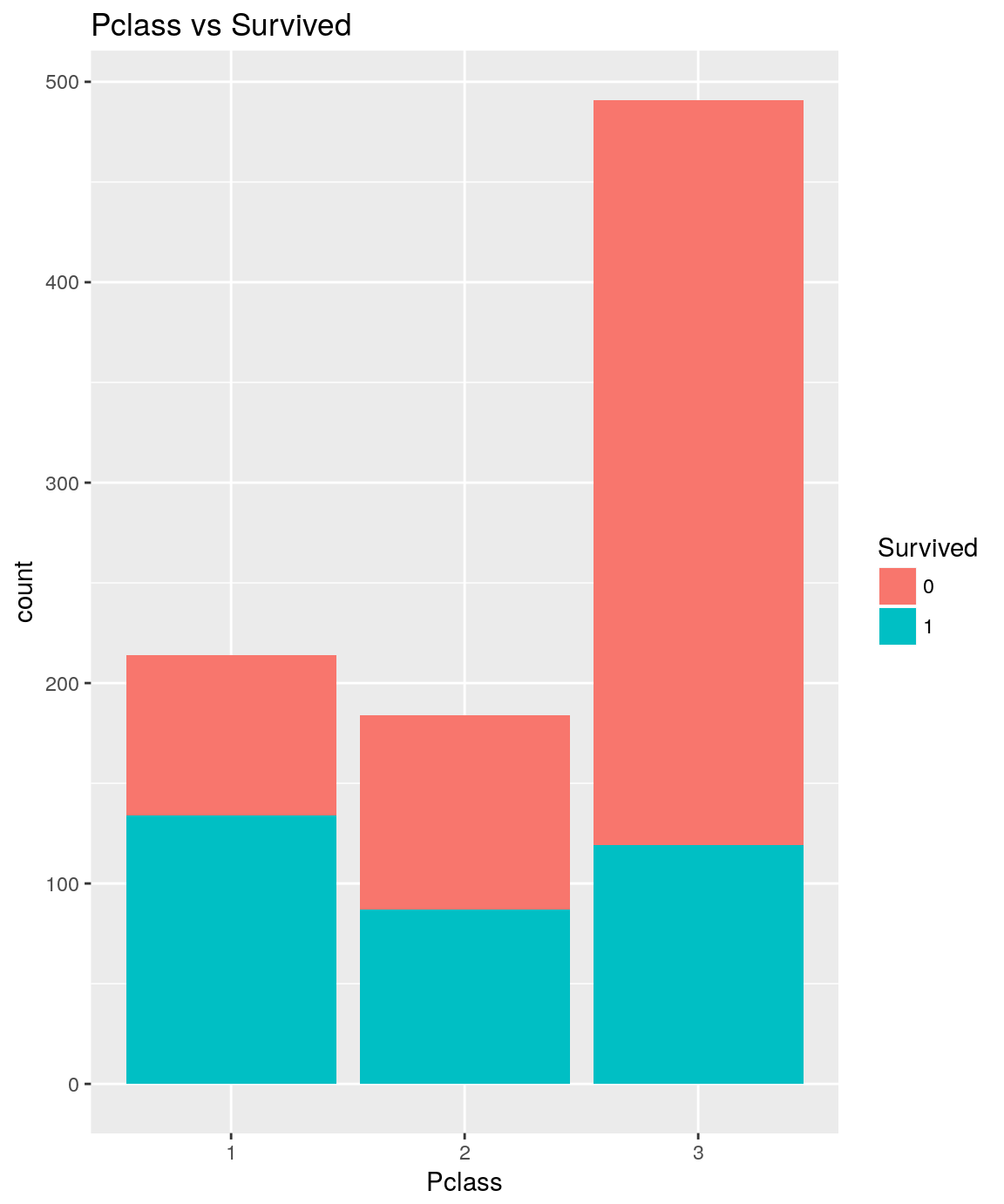
titanicTrainingDataset %>%
group_by(Pclass) %>%
summarize(SurvivedRatio = mean(Survived == 1))
## # A tibble: 3 x 2
## Pclass SurvivedRatio
## <fct> <dbl>
## 1 1 0.626
## 2 2 0.473
## 3 3 0.242
The more the passenger belongs to a wealthy class, more its likelihood to survive is high.
SibSp vs Survived
analysePredictorResponse(titanicTrainingDataset, 'SibSp', 'Survived')
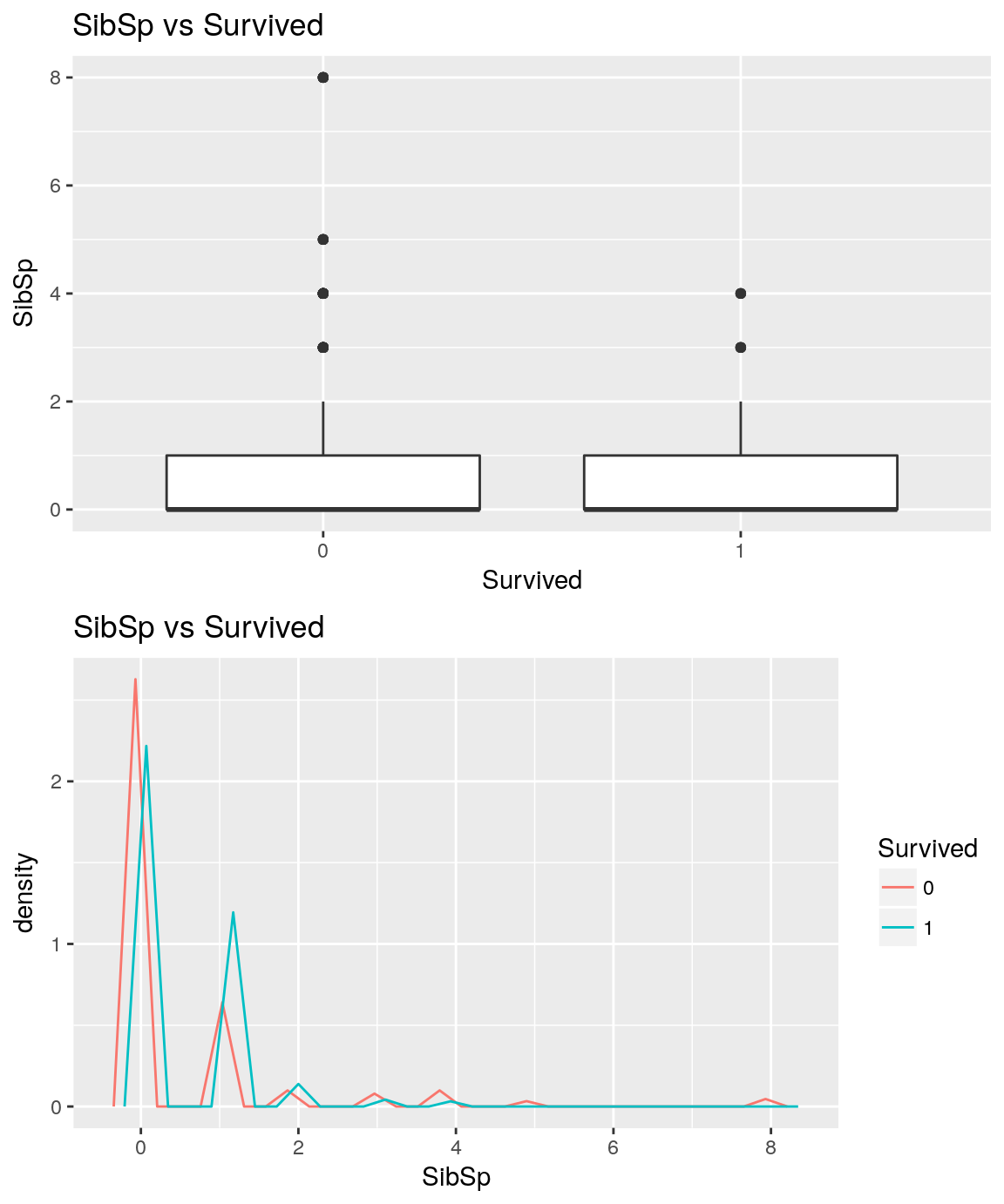
The one who have no sibling seems to have higher chance to die than the one who have one sibling onboard.
Parch vs Survived
analysePredictorResponse(titanicTrainingDataset, 'Parch', 'Survived')
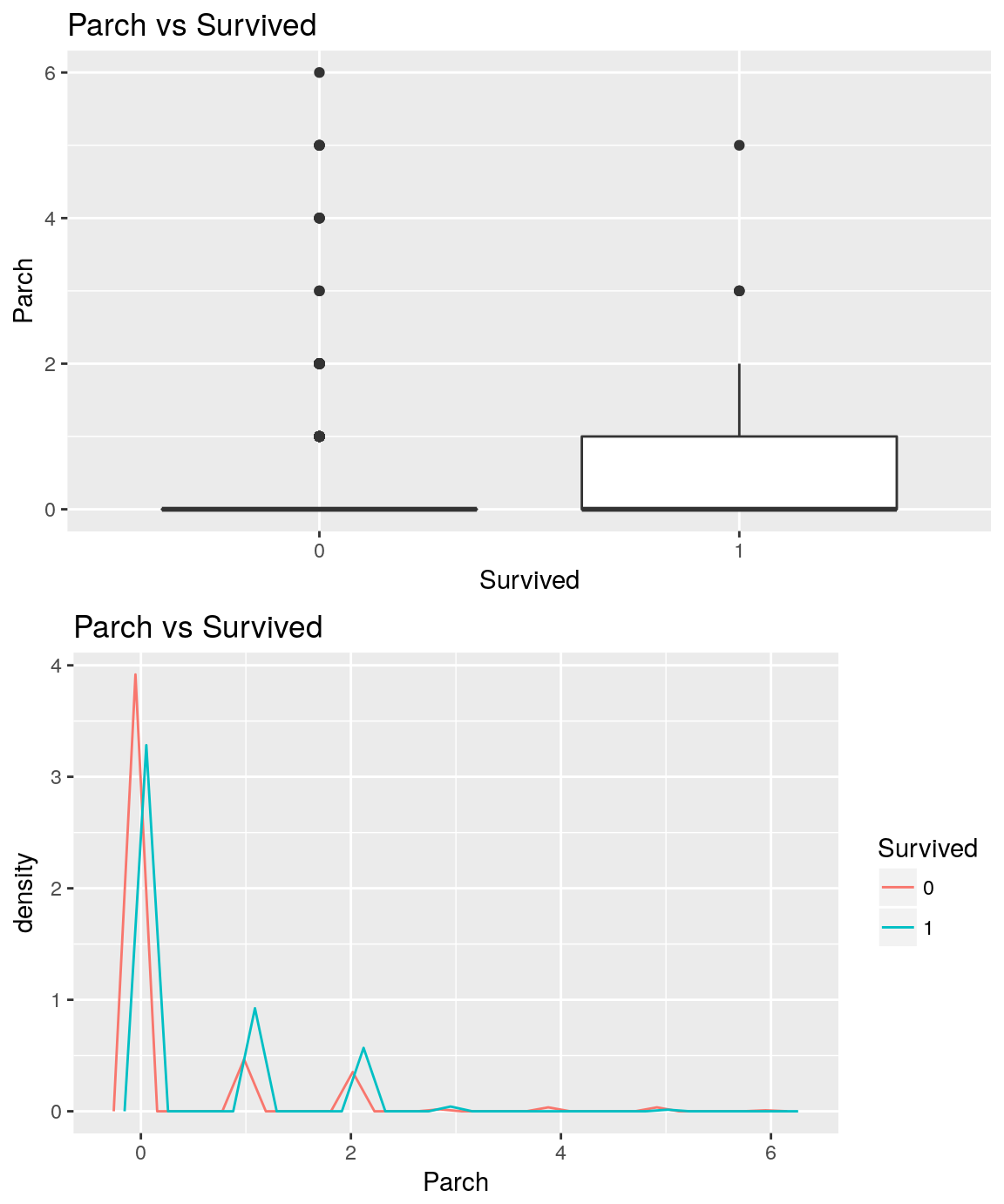
Similarly to SibSp, the one who have no parent nor children onboard seems to have higher chance to die than the one who have one parent or children.
Fare vs Survived
analysePredictorResponse(titanicTrainingDataset, 'Fare', 'Survived')
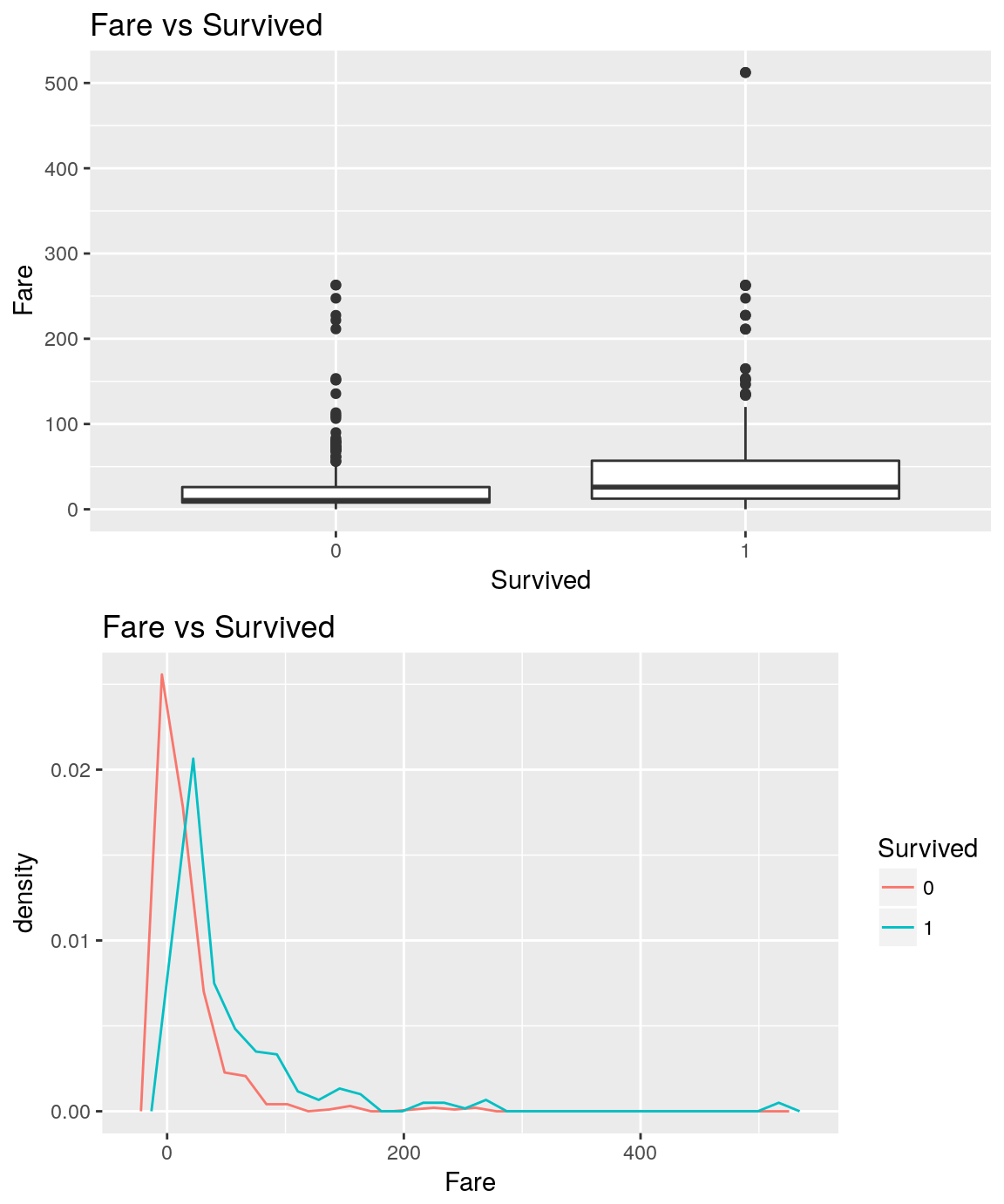
On average, higher is the fare, higher seems the likelihood to survive.
Embarked vs Survived
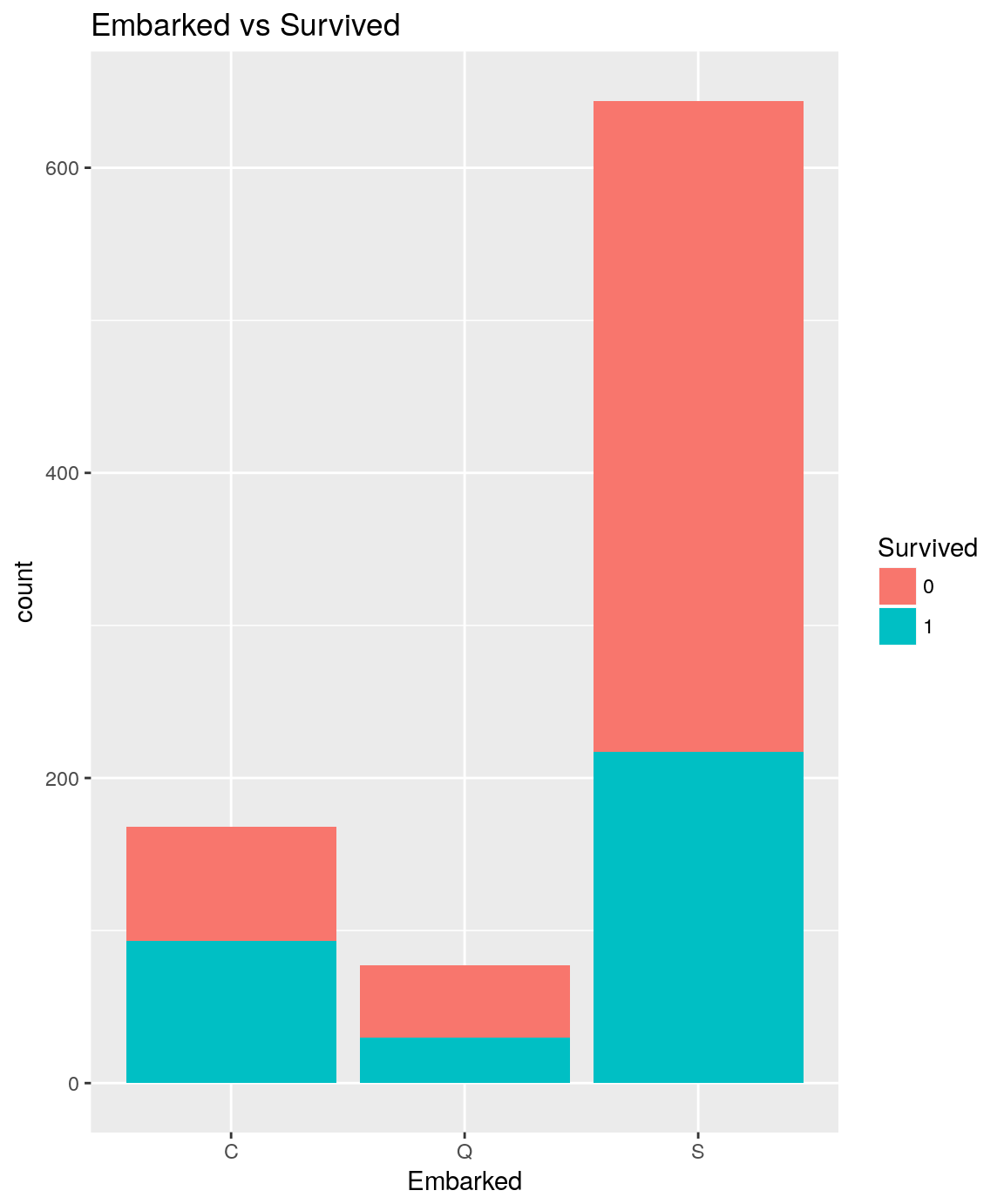
analysePredictorResponse(titanicTrainingDataset, 'Embarked', 'Survived')
titanicTrainingDataset %>%
group_by(Embarked) %>%
summarize(SurvivedRatio = mean(Survived == 1))
## # A tibble: 3 x 2
## Embarked SurvivedRatio
## <fct> <dbl>
## 1 C 0.554
## 2 Q 0.390
## 3 S 0.337
It seems there are some significant differences regarding the Survived rate depending on the port of Embarkation. Port C lead to 55.36% survive rate whereas port S lead to only 33.69%.
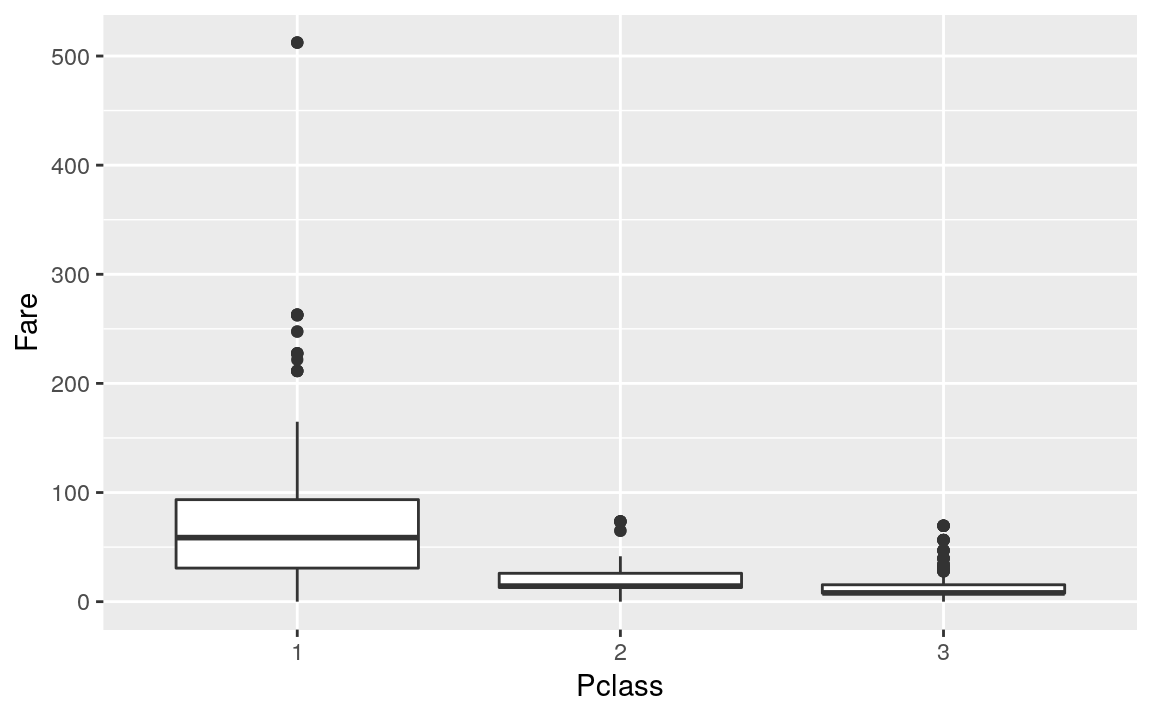
Pclass vs Fare
titanicTrainingDataset %>%
ggplot(mapping = aes(Pclass, Fare)) +
geom_boxplot()
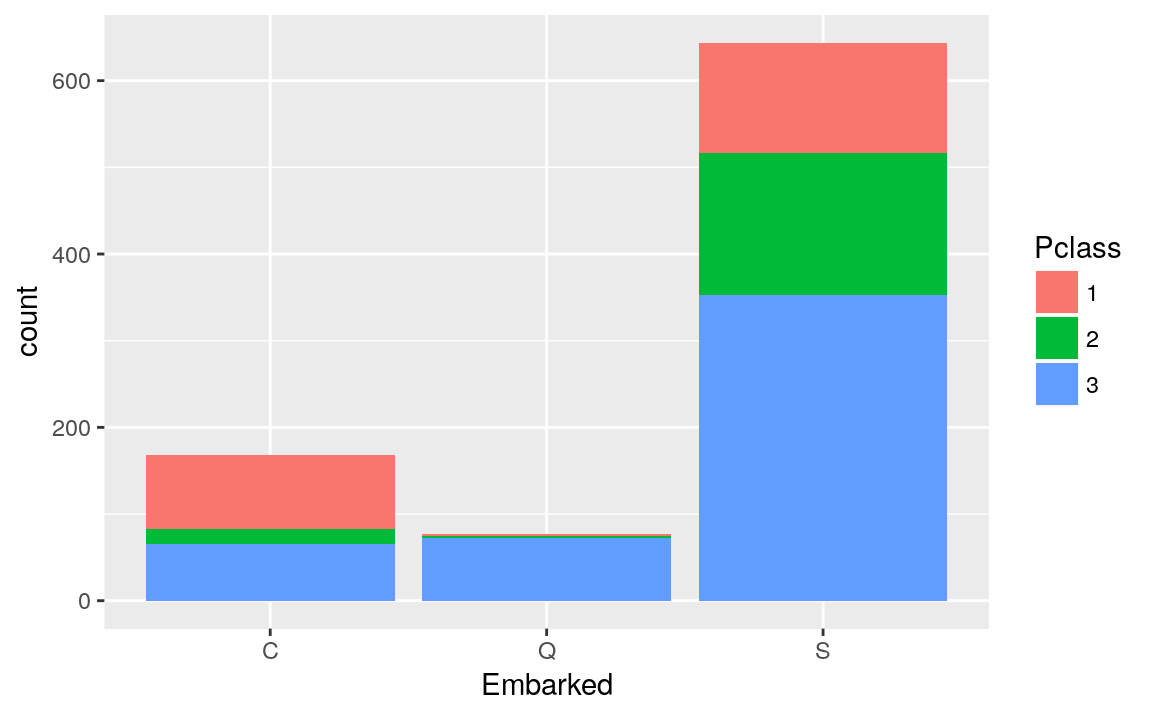
** Embarked vs Pclass**
titanicTrainingDataset %>%
ggplot(mapping = aes(Embarked, fill = Pclass)) +
geom_bar()
What about the variable Title that I have extracted from the names ?
titanicTrainingDataset %>%
group_by(Title) %>%
summarize(SurvivedRatio = mean(Survived == 1)) %>%
arrange(SurvivedRatio) %>%
mutate(Title = factor(Title, levels = Title)) %>%
ggplot(aes(x = Title, y = SurvivedRatio)) +
geom_col() +
coord_flip()
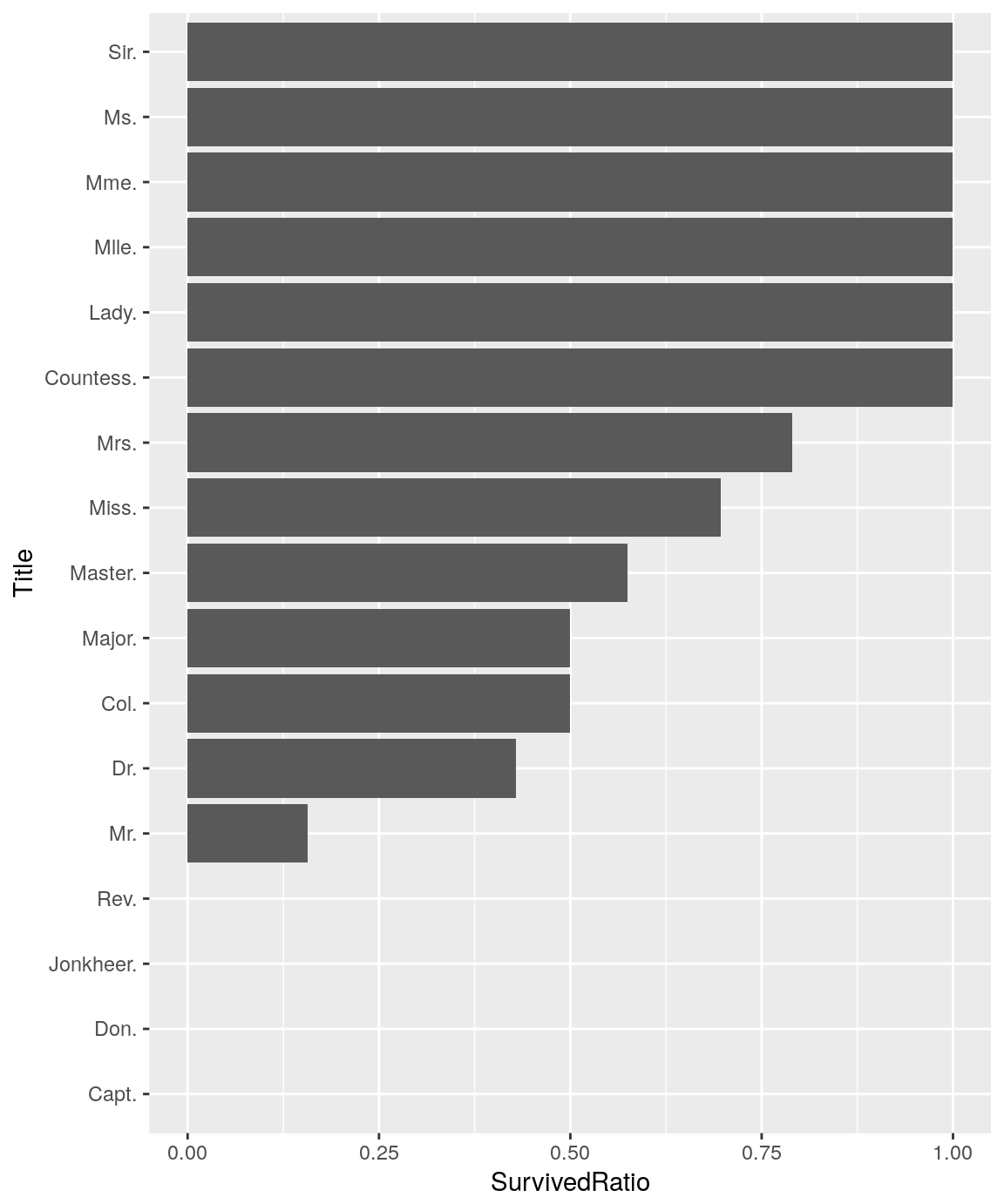
Passengers title seems to provide interresting information for predicting the surviving ones. It seems we can refine this variable in order to reduce the number of levels. I Added a new categorical variable 'RefinedTitle' that split the passengers titles into 3 relevant levels :
titanicTrainingDataset <- titanicTrainingDataset %>% mutate(
RefinedTitle = factor(ifelse(Title %in% c('Capt.', 'Don', 'Jonkheer.', 'Rev.', 'Mr.'), 1,
ifelse(Title %in% c('Col.', 'Dr.', 'Major.', 'Master.'), 2, 3
)
))
)
Modelisation
Logistic regression
Here is a first attempt of modelisation on the training dataset with most of the candidate predictors :
model <- glm(Survived ~ RefinedTitle + Pclass + SibSp + Embarked + Age + Sex + Parch, data = titanicTrainingDataset %>% filter(!is.na(Age)), family = binomial)
summary(model)
##
## Call:
## glm(formula = Survived ~ RefinedTitle + Pclass + SibSp + Embarked +
## Age + Sex + Parch, family = binomial, data = titanicTrainingDataset %>%
## filter(!is.na(Age)))
##
## Deviance Residuals:
## Min 1Q Median 3Q Max
## -2.3632 -0.5951 -0.3917 0.5812 2.5346
##
## Coefficients:
## Estimate Std. Error z value Pr(>|z|)
## (Intercept) 0.930213 0.393458 2.364 0.018069 *
## RefinedTitle2 2.556167 0.426595 5.992 2.07e-09 ***
## RefinedTitle3 0.473372 1.439032 0.329 0.742192
## Pclass2 -1.032744 0.282392 -3.657 0.000255 ***
## Pclass3 -2.163955 0.262944 -8.230 < 2e-16 ***
## SibSp -0.452722 0.114388 -3.958 7.56e-05 ***
## EmbarkedQ -0.274539 0.392170 -0.700 0.483895
## EmbarkedS -0.484066 0.244469 -1.980 0.047696 *
## Age -0.027045 0.008209 -3.294 0.000986 ***
## Sexfemale 2.672175 1.448318 1.845 0.065035 .
## Parch -0.189686 0.122467 -1.549 0.121413
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## (Dispersion parameter for binomial family taken to be 1)
##
## Null deviance: 1182.8 on 888 degrees of freedom
## Residual deviance: 745.7 on 878 degrees of freedom
## AIC: 767.7
##
## Number of Fisher Scoring iterations: 5
Parch and Embarked don't seem to be relevant according to P-value associated with the Z-statistic. Also, Sex seems to be confounding with RefinedTitle for predicting 'Survived'. Here is a refined model without Parch, Embarked and Sex variables :
model <- glm(Survived ~ RefinedTitle + Pclass + SibSp + Age, data = titanicTrainingDataset, family = binomial)
summary(model)
##
## Call:
## glm(formula = Survived ~ RefinedTitle + Pclass + SibSp + Age,
## family = binomial, data = titanicTrainingDataset)
##
## Deviance Residuals:
## Min 1Q Median 3Q Max
## -2.5200 -0.5798 -0.3934 0.5919 2.6299
##
## Coefficients:
## Estimate Std. Error z value Pr(>|z|)
## (Intercept) 0.646841 0.368287 1.756 0.079029 .
## RefinedTitle2 2.484859 0.415967 5.974 2.32e-09 ***
## RefinedTitle3 3.066697 0.207374 14.788 < 2e-16 ***
## Pclass2 -1.153534 0.271979 -4.241 2.22e-05 ***
## Pclass3 -2.227890 0.250689 -8.887 < 2e-16 ***
## SibSp -0.524237 0.110349 -4.751 2.03e-06 ***
## Age -0.028450 0.008128 -3.500 0.000465 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## (Dispersion parameter for binomial family taken to be 1)
##
## Null deviance: 1182.82 on 888 degrees of freedom
## Residual deviance: 754.59 on 882 degrees of freedom
## AIC: 768.59
##
## Number of Fisher Scoring iterations: 5
All the coefficient estimates for predictors are now statistically significant according to their P-Values.
Estimate the test error rate of the logistic regression with LOOCV
Here is a perform of a "leave-one-out" cross validation over several decision boundary values (from 0.1 to 0.9) in order to find the value that minimize the simulated test error rate with the training dataset. For each value, it will display the estimated error rate along with the confusion matrix.
for (j in seq(.1, .9, .1)) {
predictions <- rep(0, nrow(titanicTrainingDataset))
for (i in 1:nrow(titanicTrainingDataset)) {
model <- glm(Survived ~ RefinedTitle + Pclass + SibSp + Age, data = titanicTrainingDataset, family = binomial, subset = -i)
predictions[i] <- predict(model, titanicTrainingDataset[i,], type = "response") > j
}
print(paste("Decision boundary value :", j))
print(table(predictions, titanicTrainingDataset$Survived))
print(mean(predictions != titanicTrainingDataset$Survived))
}
## [1] "Decision boundary value : 0.1"
##
## predictions 0 1
## 0 250 33
## 1 299 307
## [1] 0.3734533
## [1] "Decision boundary value : 0.2"
##
## predictions 0 1
## 0 346 44
## 1 203 296
## [1] 0.2778403
## [1] "Decision boundary value : 0.3"
##
## predictions 0 1
## 0 423 57
## 1 126 283
## [1] 0.2058493
## [1] "Decision boundary value : 0.4"
##
## predictions 0 1
## 0 443 68
## 1 106 272
## [1] 0.1957255
## [1] "Decision boundary value : 0.5"
##
## predictions 0 1
## 0 482 92
## 1 67 248
## [1] 0.1788526
## [1] "Decision boundary value : 0.6"
##
## predictions 0 1
## 0 501 110
## 1 48 230
## [1] 0.1777278
## [1] "Decision boundary value : 0.7"
##
## predictions 0 1
## 0 523 152
## 1 26 188
## [1] 0.200225
## [1] "Decision boundary value : 0.8"
##
## predictions 0 1
## 0 538 198
## 1 11 142
## [1] 0.2350956
## [1] "Decision boundary value : 0.9"
##
## predictions 0 1
## 0 544 276
## 1 5 64
## [1] 0.3160855
Estimated test error rate seems to be minimized with a decision boundary values of 0.6 resulting a test error rate of 17.77%.
Use the logistic regression model to predict the Survived variable with the test dataset
model <- glm(Survived ~ RefinedTitle + Pclass + SibSp + Age, data = titanicTrainingDataset, family = binomial)
titanicTestDataset <- read_csv(
testFile,
col_types = cols(
Pclass = col_factor(1:3),
Sex = col_factor(c("male", "female")),
Embarked = col_factor(c("C", "Q", "S"))
)
) %>%
mutate(Title = as.factor(str_extract(Name, regex("([a-z]+\\.)", ignore_case = T)))) %>%
mutate(
RefinedTitle = factor(ifelse(Title %in% c('Capt.', 'Don', 'Jonkheer.', 'Rev.', 'Mr.'), 1,
ifelse(Title %in% c('Col.', 'Dr.', 'Major.', 'Master.'), 2, 3
)
))
) %>%
mutate(Age = ifelse(is.na(Age), median(Age, na.rm = T), Age))
predictions <- predict(model, titanicTestDataset, type = "response") > .6
tibble(PassengerId = titanicTestDataset$PassengerId, Survived = as.integer(predictions)) %>%
write_csv('predictions-logistic-regression.csv')
Kaggle score is 77.5% with this model.
Quadratic Discriminant Analysis
model <- qda(Survived ~ RefinedTitle + Pclass + SibSp + Age, data = titanicTrainingDataset, family = binomial)
model
## Call:
## qda(Survived ~ RefinedTitle + Pclass + SibSp + Age, data = titanicTrainingDataset,
## family = binomial)
##
## Prior probabilities of groups:
## 0 1
## 0.6175478 0.3824522
##
## Group means:
## RefinedTitle2 RefinedTitle3 Pclass2 Pclass3 SibSp Age
## 0 0.04189435 0.1493625 0.1766849 0.6775956 0.5537341 30.02823
## 1 0.08235294 0.6794118 0.2558824 0.3500000 0.4764706 28.16374
LOOCV on the QDA model
predictions <- factor(rep(0, nrow(titanicTrainingDataset)), levels = 0:1)
for (i in 1:nrow(titanicTrainingDataset)) {
model <- qda(Survived ~ RefinedTitle + Pclass + SibSp + Age, data = titanicTrainingDataset, subset = -i)
predictions[i] <- predict(model, titanicTrainingDataset[i,])$class
}
table(predictions, titanicTrainingDataset$Survived)
##
## predictions 0 1
## 0 451 78
## 1 98 262
mean(predictions != titanicTrainingDataset$Survived)
## [1] 0.1979753
Estimated test error rate is 19.79% with the Quadratic Discriminant Analysis model.
Use the QDA model to predict the Survived variable with the test dataset from kaggle
model <- qda(Survived ~ RefinedTitle + Pclass + SibSp + Age, data = titanicTrainingDataset)
predictions <- predict(model, titanicTestDataset)$class
tibble(PassengerId = titanicTestDataset$PassengerId, Survived = predictions) %>%
write_csv('predictions-qda.csv')
Kaggle score is 77.5%, almost like the logistic regression.
K-nearest neighbors
Lets keep the same set of predictors that seems to be relevant to predict the response, and use the KNN algorithm with them. First, I evalute the best K value with a Validation Set approach, by splitting the training dataset into a training and a test datasets :
set.seed(1)
titanicTrainingDatasetKnn <- titanicTrainingDataset
titanicTrainingDatasetKnn$RefinedTitle = as.integer(titanicTrainingDatasetKnn$RefinedTitle)
titanicTrainingDatasetKnn$Pclass = as.integer(titanicTrainingDatasetKnn$Pclass)
testSampleSize <- 150
isTest <- sample(nrow(titanicTrainingDataset), testSampleSize)
knnTrainDataset <- titanicTrainingDatasetKnn[-isTest,] %>%
subset(select = c(RefinedTitle, Pclass, SibSp, Age)) %>%
scale()
knnTestDataset <- titanicTrainingDatasetKnn[isTest,] %>%
subset(select = c(RefinedTitle, Pclass, SibSp, Age)) %>%
scale()
cl <- titanicTrainingDatasetKnn[-isTest,]$Survived
errorsRate <- rep(0, testSampleSize)
for (k in 1:testSampleSize) {
predictions <- knn(knnTrainDataset, knnTestDataset, cl, k)
errorsRate[k] = mean(predictions != titanicTrainingDatasetKnn[isTest,]$Survived)
}
tibble(k = 1:testSampleSize, errorsRate = errorsRate) %>%
ggplot(aes(x = k, y = errorsRate)) +
geom_path()
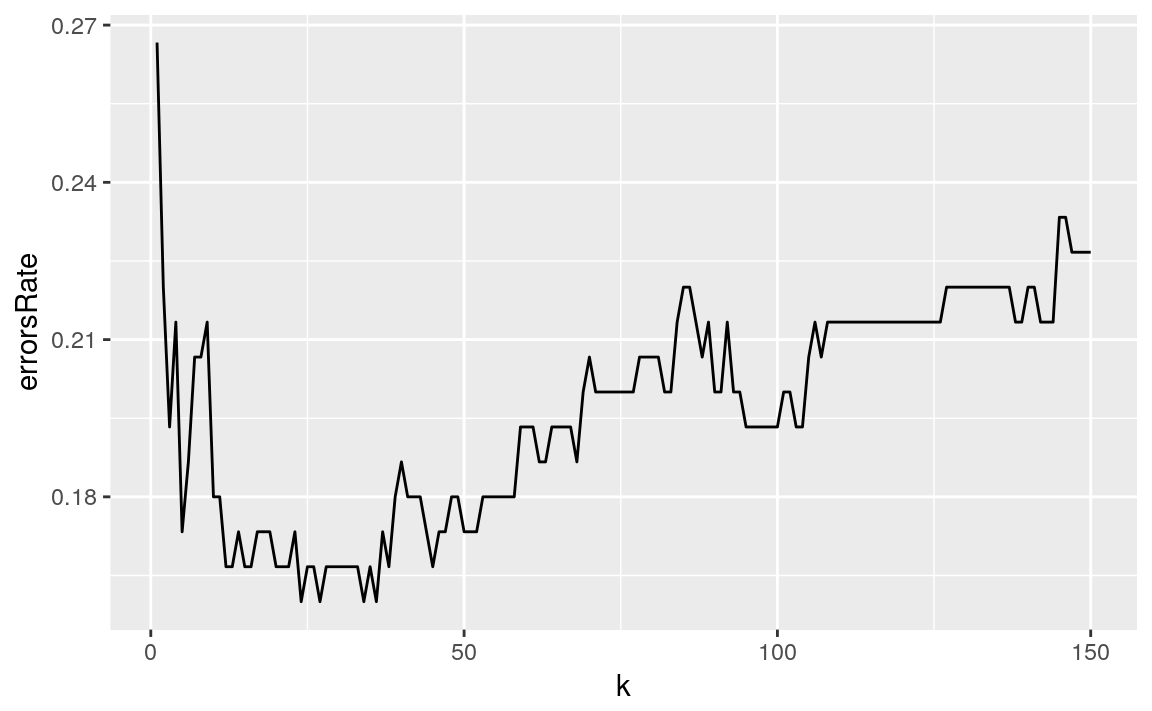
I Choose k = 35 for minimizing the test error rate.
Use KNN (K = 35) to predict the response with the test dataset
titanicTestDatasetKnn <- titanicTestDataset %>%
mutate(RefinedTitle = as.integer(RefinedTitle)) %>%
mutate(Pclass = as.integer(Pclass))
knnTrainDataset <- titanicTrainingDatasetKnn %>%
subset(select = c(RefinedTitle, Pclass, SibSp, Age)) %>%
scale()
knnTestDataset <- titanicTestDatasetKnn %>%
subset(select = c(RefinedTitle, Pclass, SibSp, Age)) %>%
scale()
cl <- titanicTrainingDatasetKnn$Survived
predictions <- knn(knnTrainDataset, knnTestDataset, cl, 35)
tibble(PassengerId = titanicTestDataset$PassengerId, Survived = predictions) %>%
write_csv('predictions-knn.csv')
Kaggle score is 78.94%.
